UM achieves new breakthrough in biochip research
澳大高效生物芯片研究再獲突破
28 Apr 2016
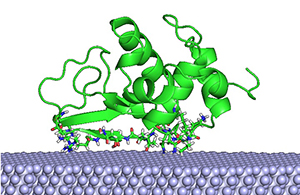 The 3D protein-surface structures of Lysozyme on the surface of PTFE
The 3D protein-surface structures of Lysozyme on the surface of PTFE溶菌酶Lysozyme在聚四氟乙烯表面的三維構像
The University of Macau (UM) has achieved a breakthrough in the research of biochips. The Computational Biology and Bioinformatics Lab (CBBio), under UM's Faculty of Science and Technology, and the State-Key Laboratory of Analog and Mixed-Signal VLSI, have successfully developed a computational intelligence-based software programme to address the challenges of conformational sampling in 3D protein-surface structure prediction, which helps to produce biochips with the ability to locate the optimal protein configuration in the most promising low-energy region. The work was recently published by Oxford University Press's journal Bioinformatics, the most authoritative publication in the field of bioinformatics and computational biology.
In order to predict the optimal protein configuration computationally, the research team translated the problem into an optimisation task, to search intelligently among all possible orientations and positions of protein with respect to the surface, for the most promising low-energy region where the optimal is. Inspired by nature, the ‘intelligent’ part of the project is based on the social behaviour model of bird flocking when searching for food. Birds in a flock would memorise places where the most food has been found, and communicate with one another to update the best food source. Based on this knowledge, the birds iteratively adjust their flying direction towards previously successful regions until no better food source is found.
By combining the search model with a newly devised forcefield-based energy function describing the protein-surface interactions and fine-tuned parameters, the research team has created an efficient computer algorithm to address the protein-surface structure prediction problem, ProtPOS. The programme (http://cbbio.cis.umac.mo/software/protpos/) can work seamlessly with most popular molecular simulation software, so prediction results can be used right away in further computational studies for biochip design.
The research project was conducted by UM PhD student Ngai Choi Fong, jointly supervised by Dr Shirley Siu Weng In and Prof Elvis Mak Pui In, and funded by UM.
澳門大學最近在生物芯片發展研究再有突破,成功設計出有效預測蛋白質表面吸附結構的智慧型計算方法,有助研發以蛋白質為配體的生物微型芯片。該研究獲相關領域中最權威的牛津大學出版社期刊《Bioinformatics》刊登。
該項研究由澳大科技學院生物信息學實驗室與模擬與混合信號超大規模集成電路國家重點實驗室進行,成功開發出以智慧型計算為本的算法,解決了蛋白質吸附在固體表面上的三維結構預測中構像取樣的問題。該算法靈感來自於大自然,建基於鳥群覓食的社會行為模式:鳥兒記住見過最多食物的地方,亦透過互相通訊來更新鳥群所知的最佳食物來源,透過這些資訊,他們會反復調整飛行的方向以朝著認為比較可行的地方前進,直到沒有找到更好的食物源為止。
澳大研究員結合上述搜索模式與描述蛋白質表面相互作用能量的函數,建立了一個有效的蛋白質表面吸附結構預測計算機算法。透過優化蛋白質構像的預測,研究員便可從所有可能的表面位置和蛋白質的吸附方向中,搜索出最低能量的蛋白質三維構像。相關程式更與最流行的分子模擬軟件合作無縫(詳見於網站http://cbbio.cis.umac.mo/software/protpos/),使預測的結果可立即用作進一步生物芯片設計中的計算研究。
